The radiology department at ZGT in the Netherlands (https://www.zgt.nl) recently migrated all of the PACS data from their current PACS to a new PACS. Previously the department had utilised Pukka-j’s DICOM products to migrate their data from their previous PACS so when the time came to move their data again, they came to Pukka-j to find the fastest way to move it. With the new Nexus software, the department was able to take greater control over their own migration process with the help of some input from the experts at Pukka-j. This article describes the process followed to successfully move to another PACS provider.
The first stage, as with all migrations was to analyse the current situation. The data was compressed as JPEG2000, with ~30Tb spread across 11 network storage locations. As with many PACSs, the DICOM information in the header was out of sync with the information in the database, in this case a SQL server cluster which the department had access to. There were also references on disk that had been deleted from the database, or the references had been changed to point at different files after a study merge process, thus leaving “orphaned” files on the storage.
In order to maximise the speed at which the data could be transferred, it made sense to maintain the JPEG2000 compression as this was also supported by the destination PACS. This meant only 30Tb of data would be transferred over the network, as opposed to approximately 100Tb if it were decompressed. It was also decided that it would be optimal to read the files directly from the file system and use Nexus to reconcile the header information with that in the database, rather than relying on a query/retrieve via the PACS, that could have impacted on the service being delivered.
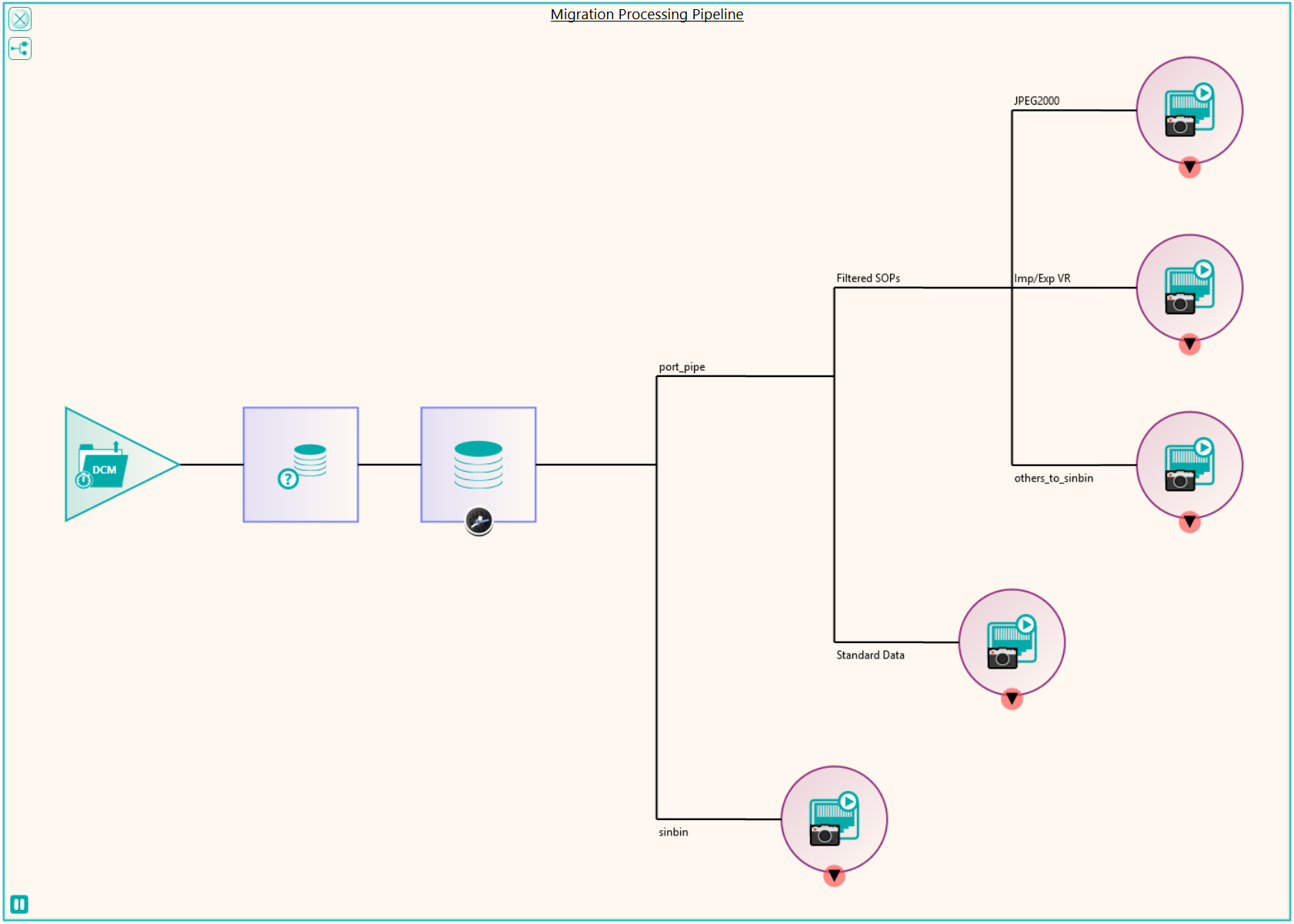
Using Nexus, it was possible to rapidly read the data using parallel processing to build a database of the contents of the file system. This highlighted that some data that contained no pixel data was stored as implicit or explicit little endian which would be required to be sent to a different port on the destination PACS. There were also some SOP classes that either weren’t supported or weren’t required on the destination PACS and would need to be filtered out. Data that also couldn’t be found in the database or couldn’t be reconciled for any other reason was also filtered out. In order to store the filtered data, Pukka-j’s Dicom Explorer PACS was set up as a “Sin Bin” archive where the department could review the filtered data and decide what needed to be done with it.
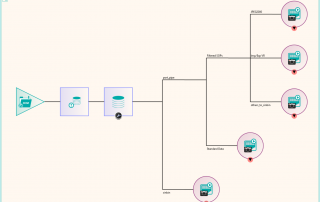
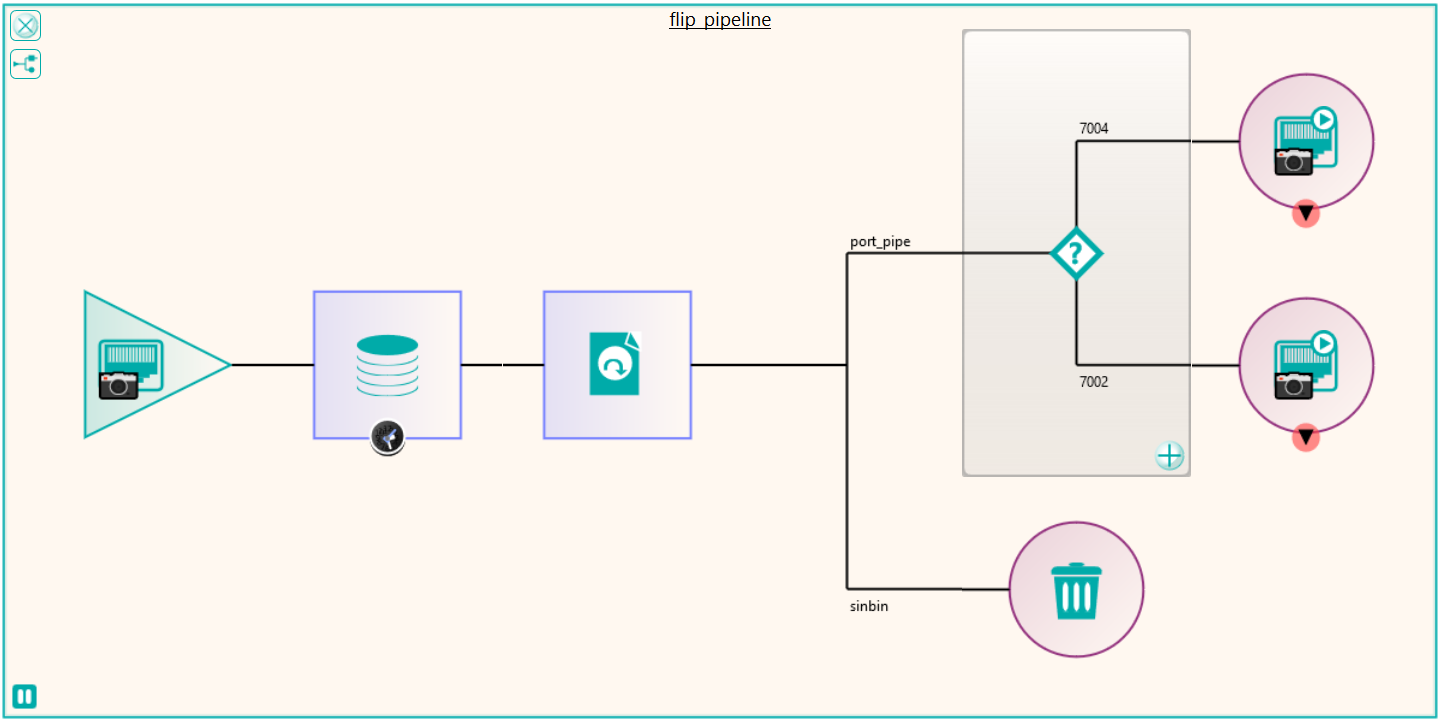
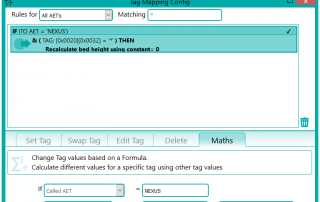
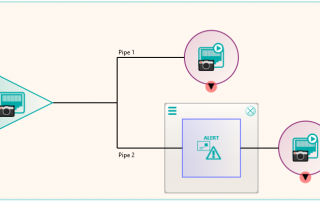
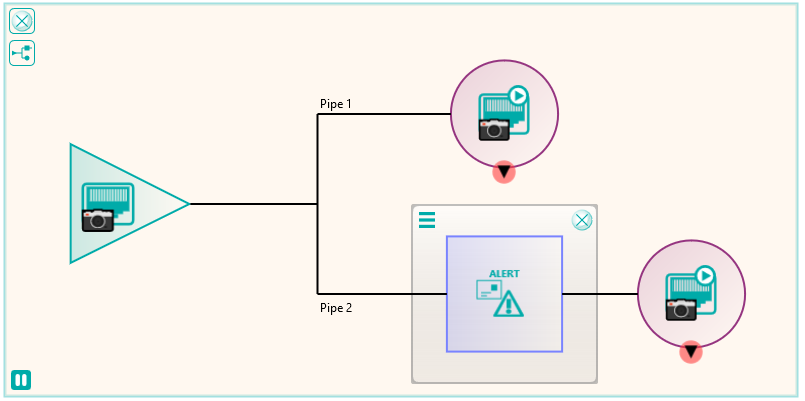
There were a number of studies that needed to be excluded from the migration process, either because they were studies that weren’t required or they would require specific processing. This was handled by the reconciliation node such that, if a study UID was identified (after reconciliation) that matched the exclusion list, it was sent to the Sin Bin. The image shows the Nexus network that was created to first read the data from the file system using multiple processing threads. This input node fed into the reconciliation node to fix up the DICOM information in the header. A database node was used after the reconciliation to keep a record of the data that had been found and whether it was to be sin-binned. The data was then filtered down the appropriate pipelines based on SOP class and transfer syntax to the appropriate DICOM port on the destination PACS.
There were a number of test runs where this pipe line was simulated by sending the data to a terminator node with and without reconciliation to get a handle on the maximum speed at which the data could first be read from the file system and then be reconciled with the database. This benchmarking process allowed the department to assess the storage capabilities of the destination PACS once the destination nodes were configured to send to the new system. Once the first trial runs of the migration were done to iron out the configuration on the destination side, some interesting effects were seen. Due to the way in which the current PACS stored studies, in which studies were “striped” across folders and due to the way the data was read by the processing threads, long delays could be seen between images in a study arriving at the destination. The destination PACS implemented a process which after a period of time, or amount of data, moved images to a slower long term archive. The result of these facts was that once delayed images were sent, then the speed at which the data could be moved started to decrease because of having to write to the slower storage. After discussion, Pukka-j were able to create a new mode of operation for reading the data from the file system, by reading the data as a stripe rather than a whole folder at time, thus minimising the time between images in a study.
Some analysis of the test runs was performed to verify the performance of the reconciliation and the routing of the data to the required ports on the PACS and after a few configuration tweaks, the migration configuration was able to be deployed across four virtual servers, each running a version of Nexus. The 11 file locations were split so each had it’s own pipe line as shown in the diagram. This allowed the processing to be done in parallel across multiple resources. When the configuration was deployed, the migration was started in earnest and ran non-stop for around 3 weeks in which time it maintained around 300,000 images per hour into the destination PACS. The whole process was able to be monitored by the department, who simply referred to Pukka-j in the event that there were any questions over any entry in the logging files.
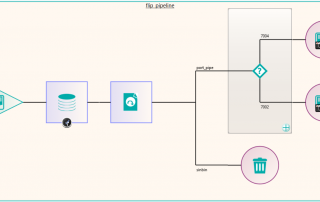
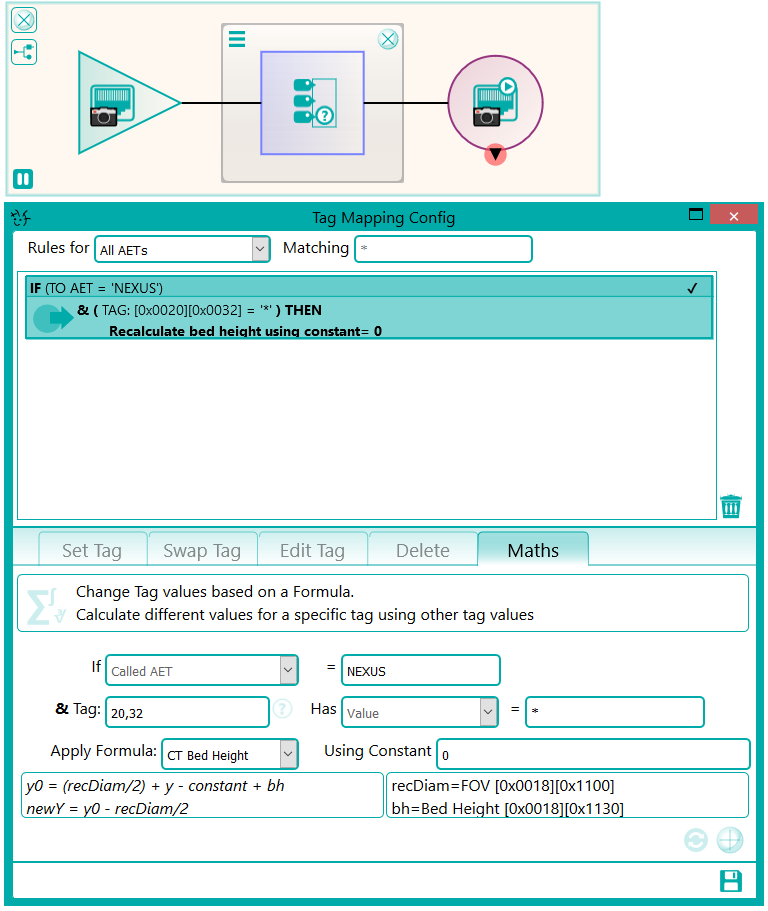
Following the migration, some of the excluded files that were not sent because they required some further processing could be dealt with. These were files that in the database had transformations recorded against them such as “rotate clockwise 90 degrees”. These were handled by a new node produced by Pukka-j to allow the department to load a list of SOP Instance UIDs and the required transformation. The data was then sent via a DICOM CMOVE through a Nexus pipeline and as they passed through the transformation node, the pixel data was modified accordingly. The network below shows the image transformation pipeline used.
Sander ten Hoeve (Functioneel Applicatiebeheerder sr) at ZGT, who managed the Nexus services throughout the migration told us:
“We decided to contact Pukka-J because of the previous successful migration and over the years we experienced that Pukka-J is able to develop software based on the needs of a client. The software is flexible, highly configurable and intuitive in use. Furthermore we have direct contact with the developer and any issues or wishes is solved and delivered very quickly.”
You can download the Nexus specification document here [Download not found]
You can see more on Nexus here